A new genetic screening technique reveals how dozens of mutations associated with autism alter the fate of developing brain cells in organoids.
“We wanted to find steps in brain development that are particularly susceptible” to autism-related gene mutations, says co-lead investigator Jürgen Knoblich, professor of synthetic biology at the Medical University of Vienna in Austria.
The approach builds off of a technique Knoblich’s research group developed in 2020 to screen genes linked to microcephaly for their ability to impair cell growth. In that study, the team used CRISPR to make an array of genetic mutations in human stem cells, grew each stem cell into an organoid and used unique genetic barcodes to track the mutations each cell accumulated.
In their new work, published in September in Nature, the team used a similar approach to generate human organoids carrying mutations in 36 genes strongly linked to autism. Each cell of the mosaic organoids contained a single mutation.
“This is the first time that I’ve seen that many genes examined all at once in the same model,” says Flora Vaccarino, professor of neuroscience at Yale University, who was not involved in this study. “It’s truly amazing.”
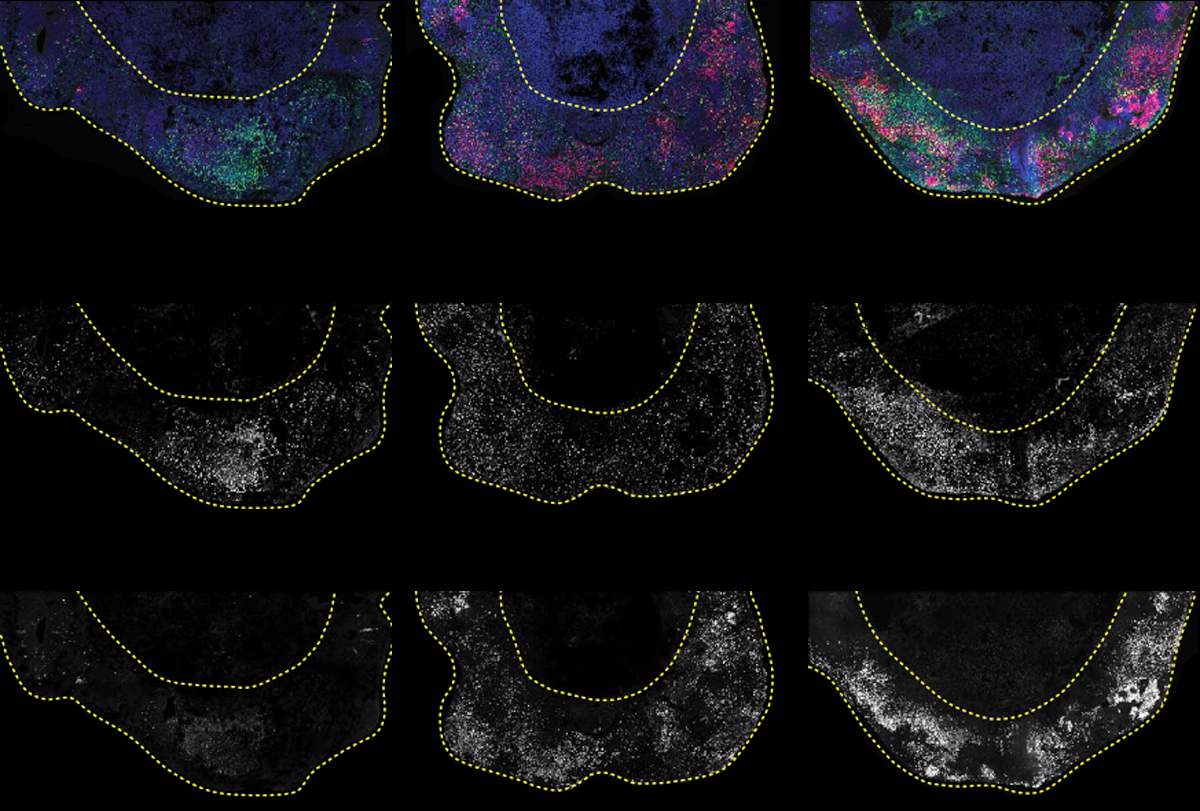
After the organoids had grown for four months and accumulated thousands of cells with each mutation, Knoblich and his colleagues used single-cell RNA sequencing to measure the abundance of individual cell types.
Cortical organoids typically grow a mix of neurons and glia arranged in layers resembling those seen in the human cerebral cortex. Mutations in 24 of the 36 genes altered the ratio of excitatory neurons to inhibitory interneurons, the researchers found. About one-third of the mutations reduced the number of intermediate progenitor cells, which give rise to excitatory neurons across cortical layers and are thought to be important for integrating information across brain regions.
“This question of imbalance seems to be a common denominator of autism spectrum disorder pathogenesis,” Vaccarino says.
Mutations in one gene in particular, ARID1B, caused one of the “strongest and most interesting” outcomes in the organoids, Knoblich says. ARID1B mutations pushed more cells than usual to become oligodendrocyte precursors, which generate the electrical insulation around neurons and have been linked to autism.
Cerebral organoids derived from two people with ARID1B mutations both showed the same atypical increase in oligodendrocyte precursors as the CRISPR-engineered organoids, the study shows. “We went from a genetic screen to recruiting patients who actually have that mutation, and I’m very proud that we could go all the way through with this,” Knoblich says.
Replicating the ARID1B findings in organoids derived from people was a first step toward validating that the study’s results translate to the human brain, Knoblich says. Future research should continue to examine more people with a range of other genetic mutations associated with autism, Vaccarinio adds.
The 36 genes Knoblich’s team focused on are all associated with transcriptional regulation, controlling how cells respond to intra- and extracellular signals. But many other autism-related genes are tied to the formation and maintenance of connections between neurons.
The new approach could not only tackle this gene set, but many others, Knoblich says. “It’s a very versatile technology. We could put any list of genes into the system and use it for any organoid.”
This content was originally published here.