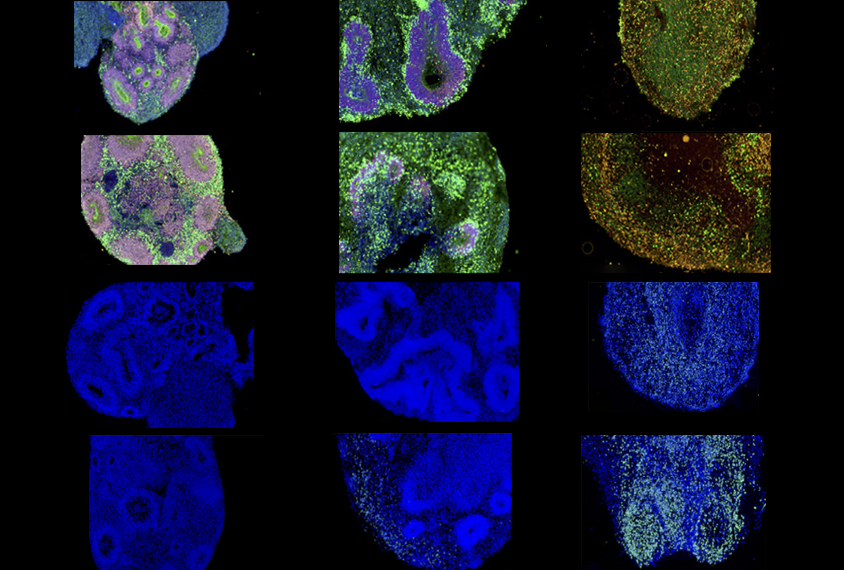
Courtesy of Shuai Fu et al. / The American Journal of Human Genetics
An autism-linked mutation disrupts the development of clusters of brain cells, or brain organoids, generated from autistic people but displays significantly less of an effect in organoids produced from neurotypical donors, a new study finds.
The study suggests that in addition to exploring the effects of known autism-linked mutations, researchers should assess the potential mediating effects of the rest of a person’s genome.
“Genetic background must be taken into consideration when studying the role of an autism spectrum disorder risk gene,” says lead investigator Anthony Wynshaw-Boris, professor of genetics and genome sciences at Case Western Reserve University in Cleveland, Ohio.
Mutations in nearly 1,000 genes are associated with autism. Mutations in the gene PTEN, which encodes a tumor-suppressing protein that also helps shape neuronal connections, account for up to 2 percent of all autism cases and up to 17 percent of those in which the child experienced brain overgrowth early in development.
Still, many people with known PTEN mutations are not autistic. This is likely because a person’s genetic background also shapes brain development. But much remains uncertain about the role of genetic background in autism, says study investigator Shuai Fu, a graduate student in Wynshaw-Boris’ lab.
In the new study, the researchers grew brain organoids from skin cells collected from two autistic boys, aged 7 and 16, and two neurotypical boys, aged 8 and 17. They developed organoids with each of these four genetic backgrounds in which all cells had either a wildtype version of PTEN or a mutated version called PTEN p.Ile135Leu. The group’s previous work linked PTEN p.Ile135Leu with autism and macrocephaly.
In both organoids with autistic and neurotypical genetic backgrounds, the p.Ile135Leu variant accelerated the proliferation of neural progenitor cells, increased the size of the organoids and modified gene activity related to neuron production, neural development and brain signaling. However, the variant led to the overproduction of both deep- and upper-layer neurons and accelerated the maturation of upper-layer neuron in organoids derived from autistic donors but not neurotypical ones. The researchers detailed their findings online 24 April in the American Journal of Human Genetics.
“A lot of research in the field focuses on specific mutations without testing for genetic background,” says Simone Mayer, an independent research group leader at the Hertie Institute for Clinical Brain Research at the University of Tübingen in Germany, who did not take part in this research. “It is great to see how the authors of this study managed to study the effect of a specific variant in combination with the genetic background.”
Even with wildtype PTEN, organoids with autistic genetic backgrounds displayed increased proliferation of neural progenitors and modified gene activity related to neuron production, neural development and brain signaling. Of the roughly 3,000 to 3,300 genes that showed either reduced or increased activity in organoids derived from autistic donors, about 90 are related to the PI3K-AKT pathway, upon which PTEN acts to influence cell growth and death.
Although PTEN p.Ile135Leu altered the activity of genes involved in neuron production and neural development in both organoids with autistic and neurotypical genetic backgrounds, the effects were at times in the opposite direction. For example, the mutation boosted the activity of genes involved in ribosome production in cells derived from neurotypical donors but reduced it in those from autistic donors.
Wynshaw-Boris plans to expand the study to include organoids from more people with autism, including women. Autism is diagnosed more often in boys and men than in girls and women, raising the possibility of a “female protective effect” linked to the X chromosome.
The scientists also want to investigate whether PTEN p.Ile135Leu and autistic genetic backgrounds affect epigenetic modifications, which can alter gene expression without changing the DNA sequence, and whether other autism-linked mutations and background variants share targets, as is the case with PTEN and PI3K-AKT. They will then test whether modifying one or more of these common targets can alter the course of neural development, Fu says.
The study findings and future work may have both diagnostic and therapeutic value, says Santhosh Girirajan, associate professor of genomics at Pennsylvania State University in State College, who was not involved in the study.
“Clinical diagnosis should not stop at primary variant discovery but should also consider looking at the genetic background,” Girirajan says. “At the therapeutic level, identifying modifiers that can alleviate effects of the primary variant would be key to finding therapeutic strategies.”
Cite this article: https://doi.org/10.53053/TRFC5698
This content was originally published here.