A forthcoming resource aims to make more than 1 million connectomes available to users of the UK Biobank, according to an unreviewed paper posted to the preprint server bioRxiv in March.
The UK Biobank contains genetic, medical, behavioral and socioeconomic data from half a million adults in the United Kingdom. About 40,000 of the participants have also completed brain scans.
Though the biobank already provides access to these scans, the images don’t contain finished data on networks and connections in the brain. That means researchers studying brain connectivity need to do additional processing, says study investigator Sina Mansour L., research fellow in psychiatry at the University of Melbourne in Australia. This introduces opportunities for variance across labs that could hinder the ability to reproduce results, Mansour says. “Even the slightest changes could have an impact on the findings.”
The connectomes will be a boon for the many labs that don’t have the storage and computational power to download and use raw biobank data, says Katherine Elizabeth Lawrence, assistant professor of research neurology at the University of Southern California in Los Angeles, who was not involved in the study but works with UK Biobank data. “If it’s already been done, they can say, ‘OK, great. Now I can use this as a starting point to answer my scientific questions.’”
The resource contains structural and functional connectivity data from 21,951 women and 19,569 men aged 44 to 82 years at the time of their first scan. Nearly 3,500 of the participants have also completed a second scan.
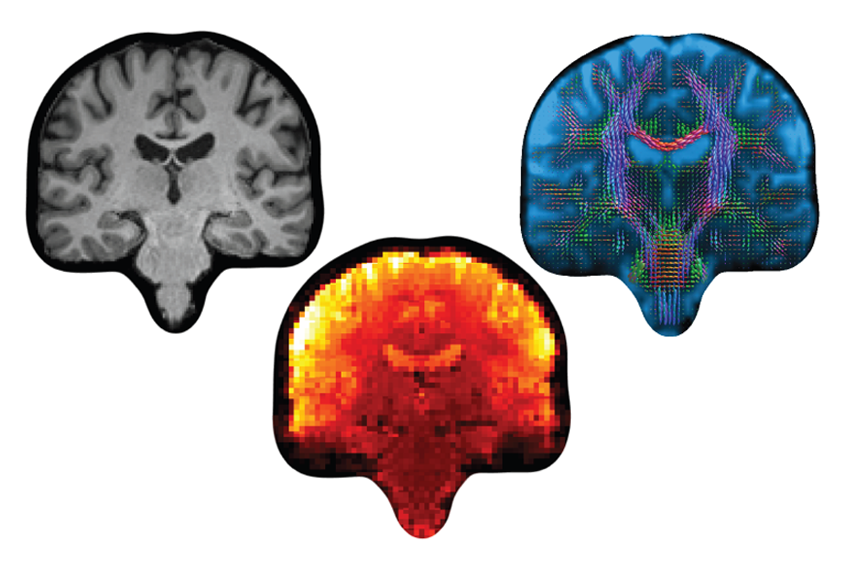
For each person in the dataset, Mansour and his team first divided the brain images into as many as 1,000 regions based on reference maps from existing datasets, including the Human Connectome Project. They then used functional MRI scans to generate networks of brain regions that activate or deactivate simultaneously.
Processing pipeline: High-performance computing clusters enabled researchers to turn UK Biobank data into downloadable maps of functional and structural connectivity.
They also made structural connectomes from diffusion MRI scans by using an algorithm to trace and quantify the white-matter connections between regions, in a process known as tractography. Altogether, the team created 27 functional and 28 structural maps per person.
The entire pipeline takes about four hours per participant. Generating the whole dataset on a single computer for all 40,000 people would take more than 20 years. But Mansour’s team used high-performance computing clusters to complete the computing and analysis in less than one year, Mansour says. For some research questions, the resulting data require just one laptop to analyze, he says.
The dataset will be useful for researchers studying sex differences in conditions such as autism, Lawrence says. It could also be used to investigate the effects of specific genetic variants on brain connectivity.
The existing UK Biobank data can also be used for those questions, but the new dataset provides a higher level of detail, particularly about the ways networks are connected, she says. “It lets you ask more questions because it lets you look at more properties of the brain.”
Because the participants are aged 44 or older, the dataset can’t reveal anything about autism earlier in life. But the data could be used to study typical changes in adult brain connectivity over time, Mansour says, and to look for variations from that in conditions such as autism or schizophrenia.
Researchers could also look for connectivity-based autism subtypes, which could help identify which treatments work best for whom, Mansour says.
Mansour and his team are working with the UK Biobank to make the connectomes available to anyone with biobank access, Mansour says. That will likely happen in conjunction with a published version of the paper, he says. They are also developing a procedure to automatically create connectomes as more participants are scanned.
This content was originally published here.